Catalogue Fitting¶
Synference has a method specifically for fitting catalogues of sources - fit_catalogue.
This is designed to be a flexible and fast way to fit your full catalogue.
It can handle:
Transforming observations to match the expected model features.
Prior predictive checks/outlier detection to remove sources that are unlikely to be well modelled.
Optional imputation of missing data.
Rapid posterior inference using trained model.
Optional SED recovery (see this notebook for more details).
Returning structured results.
[1]:
import os
import numpy as np
from IPython.display import display
from synthesizer import get_grids_dir
from unyt import Jy
from synference import SBI_Fitter, load_unc_model_from_hdf5, test_data_dir
print(get_grids_dir())
available_grids = os.listdir(get_grids_dir())
if "test_grid.hdf5" not in available_grids:
cmd = f"synthesizer-download --test-grids --destination {get_grids_dir()}"
os.system(cmd)
library_path = os.path.join(get_grids_dir(), "test_grid.hdf5")
/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
/home/runner/.local/share/Synthesizer/grids
We’ll use a subset of the JADES spectroscopic catalogue used in the synference paper for this example.
[2]:
from astropy.table import Table
cat = Table.read(f"{test_data_dir}/jades_spec_catalogue_subset.fits")
The first step is the same as our SED recovery example - we will load our trained model and the noise model used to train the model. We need the noise model to transform our observations to match the model features.
[3]:
library_path = (
f"{test_data_dir}/grid_BPASS_Chab_DenseBasis_SFH_0.01_z_14_logN_2.7_Calzetti_v3_multinode.hdf5" # noqa: E501
)
fitter = SBI_Fitter.load_saved_model(
model_file=f"{test_data_dir}", library_path=library_path, device="cpu"
)
nm_path = f"{test_data_dir}/BPASS_DenseBasis_v4_final_nsf_0_params_empirical_noise_models.h5"
noise_models = load_unc_model_from_hdf5(nm_path)
fitter.feature_array_flags["empirical_noise_models"] = noise_models
2025-11-13 20:09:59,354 | synference | INFO | Loaded model from /home/runner/.local/share/Synthesizer/data/synference/BPASS_DenseBasis_v4_final_nsf_0_posterior.pkl.
2025-11-13 20:09:59,355 | synference | INFO | Device: cpu
2025-11-13 20:09:59,398 | synference | WARNING | IndexError when trying to set train/test arrays.
Now we need to explain our catalogue to synference. We create a ‘conversion_dict’ dictionary which maps the feature names to columns in the table. The expected column names for flux are the SVO format filter names, e.g. JWST/NIRCam.F070W, or just F070W if it is unambiguous. For flux errors it is the same, but with ‘unc_’ prefixed, e.g. unc_JWST/NIRCam.F070W or unc_F070W. Any other features (e.g. redshift) should be mapped to the column name in the catalogue.
In this case our catalogue is almost in the correct format, so we just need to convert the band names to include the facility and instrument. We do this by getting the band names from the fitted model (fitter.feature_names) and replacing the relevant parts of the strings.
We then create our conversion dictionary to map from e.g. ‘unc_F200W’ to ‘unc_JWST/NIRCam.F200W’.
[4]:
def band_to_instrument(band):
"""Band name to instrument mapping."""
if band in ["F435W", "F606W", "F775W", "F814W", "F850LP"]:
return f"HST/ACS_WFC.{band}"
return f"JWST/NIRCam.{band}"
bands = [
i.split(".")[-1]
for i in fitter.feature_names
if not (i.startswith("unc_") or i.startswith("redshift"))
]
print(bands)
conversion_dict = {band: band_to_instrument(band) for band in bands}
conversion_dict.update({f"unc_{band}": f"unc_{band_to_instrument(band)}" for band in bands})
conversion_dict["redshift"] = "redshift"
conversion_dict
['F435W', 'F606W', 'F775W', 'F814W', 'F850LP', 'F090W', 'F115W', 'F150W', 'F200W', 'F277W', 'F335M', 'F356W', 'F410M', 'F444W']
[4]:
{'F435W': 'HST/ACS_WFC.F435W',
'F606W': 'HST/ACS_WFC.F606W',
'F775W': 'HST/ACS_WFC.F775W',
'F814W': 'HST/ACS_WFC.F814W',
'F850LP': 'HST/ACS_WFC.F850LP',
'F090W': 'JWST/NIRCam.F090W',
'F115W': 'JWST/NIRCam.F115W',
'F150W': 'JWST/NIRCam.F150W',
'F200W': 'JWST/NIRCam.F200W',
'F277W': 'JWST/NIRCam.F277W',
'F335M': 'JWST/NIRCam.F335M',
'F356W': 'JWST/NIRCam.F356W',
'F410M': 'JWST/NIRCam.F410M',
'F444W': 'JWST/NIRCam.F444W',
'unc_F435W': 'unc_HST/ACS_WFC.F435W',
'unc_F606W': 'unc_HST/ACS_WFC.F606W',
'unc_F775W': 'unc_HST/ACS_WFC.F775W',
'unc_F814W': 'unc_HST/ACS_WFC.F814W',
'unc_F850LP': 'unc_HST/ACS_WFC.F850LP',
'unc_F090W': 'unc_JWST/NIRCam.F090W',
'unc_F115W': 'unc_JWST/NIRCam.F115W',
'unc_F150W': 'unc_JWST/NIRCam.F150W',
'unc_F200W': 'unc_JWST/NIRCam.F200W',
'unc_F277W': 'unc_JWST/NIRCam.F277W',
'unc_F335M': 'unc_JWST/NIRCam.F335M',
'unc_F356W': 'unc_JWST/NIRCam.F356W',
'unc_F410M': 'unc_JWST/NIRCam.F410M',
'unc_F444W': 'unc_JWST/NIRCam.F444W',
'redshift': 'redshift'}
Now we can run the inference. We specify the catalogue table, the conversion dictionary, the input flux units (Jy), and then we have some choices.
We can choose to:
Run predictive checks to remove outliers.
Impute or remove missing data.
Run posterior inference, setting the number of posterior samples to draw.
Recover SEDs for each source.
[5]:
fitter.recreate_simulator_from_library(
override_library_path=library_path, override_grid_path="test_grid.hdf5"
)
post_tab = fitter.fit_catalogue(
cat,
columns_to_feature_names=conversion_dict,
flux_units=Jy,
check_out_of_distribution=False,
recover_SEDs=False,
missing_data_flag=np.nan,
num_samples=300,
append_to_input=False,
)
2025-11-13 20:09:59,444 | synference | INFO | Overriding internal library name from provided file path.
2025-11-13 20:09:59,708 | synference | WARNING | Failed to load cosmology from HDF5. Using Planck18 instead.
2025-11-13 20:10:09,213 | synference | INFO | Updated filters: ['HST/ACS_WFC.F435W', 'HST/ACS_WFC.F606W', 'HST/ACS_WFC.F775W', 'HST/ACS_WFC.F814W', 'HST/ACS_WFC.F850LP', 'JWST/NIRCam.F090W', 'JWST/NIRCam.F115W', 'JWST/NIRCam.F150W', 'JWST/NIRCam.F200W', 'JWST/NIRCam.F277W', 'JWST/NIRCam.F335M', 'JWST/NIRCam.F356W', 'JWST/NIRCam.F410M', 'JWST/NIRCam.F444W']
2025-11-13 20:10:09,216 | synference | INFO | Simulator recreated from library at /home/runner/.local/share/Synthesizer/data/synference/grid_BPASS_Chab_DenseBasis_SFH_0.01_z_14_logN_2.7_Calzetti_v3_multinode.hdf5.
2025-11-13 20:10:09,217 | synference | INFO | Auto applying inverse log10 transform for log10_Av.
2025-11-13 20:10:09,217 | synference | INFO | Auto applying inverse log10 transform for log10_mass_weighted_age.
2025-11-13 20:10:09,218 | synference | INFO | Auto applying inverse log10 transform for log10_floor_sfr_10.
2025-11-13 20:10:09,218 | synference | INFO | Adding Av to tau_v transform.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
2025-11-13 20:10:09,229 | synference | INFO | Removing 54 observations with missing data.
Sampling from posterior: 100%|██████████| 76/76 [00:04<00:00, 16.56it/s]
2025-11-13 20:10:13,822 | synference | INFO | Obtained posterior samples.
We can look at the output table, which is also an astropy.table.Table.
[6]:
post_tab
[6]:
| ID | log_mass_16 | log_mass_50 | log_mass_84 | log10metallicity_16 | log10metallicity_50 | log10metallicity_84 | log10_Av_16 | log10_Av_50 | log10_Av_84 | log_sfr_16 | log_sfr_50 | log_sfr_84 | sfh_quantile_25_16 | sfh_quantile_25_50 | sfh_quantile_25_84 | sfh_quantile_50_16 | sfh_quantile_50_50 | sfh_quantile_50_84 | sfh_quantile_75_16 | sfh_quantile_75_50 | sfh_quantile_75_84 | log10_mass_weighted_age_16 | log10_mass_weighted_age_50 | log10_mass_weighted_age_84 | log10_floor_sfr_10_16 | log10_floor_sfr_10_50 | log10_floor_sfr_10_84 | log_surviving_mass_16 | log_surviving_mass_50 | log_surviving_mass_84 | beta_16 | beta_50 | beta_84 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| int64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 |
| 1 | 9.938182258605957 | 10.083813190460205 | 10.243552284240723 | -3.9550020122528076 | -3.828235387802124 | -3.565597581863403 | -0.3131781733036041 | -0.184641033411026 | -0.06918044209480285 | 0.16693461477756505 | 0.51863232254982 | 0.7621653127670288 | 0.06947257131338119 | 0.20531394332647324 | 0.44698704361915587 | 0.3039834260940552 | 0.5464377701282501 | 0.7698792672157287 | 0.6327711796760559 | 0.8851063847541809 | 0.9623370385169983 | 3.189284715652466 | 3.3818153142929077 | 3.506747884750366 | 0.17403054773807528 | 0.5106507539749146 | 0.7569806981086731 | 9.706016235351562 | 9.827853202819824 | 9.96968864440918 | -1.4129000425338745 | -1.2808820009231567 | -1.1722686290740967 |
| 2 | 9.16947898864746 | 9.380227088928223 | 9.533154487609863 | -3.9187659072875975 | -3.7546868324279785 | -3.4933716106414794 | -2.943922929763794 | -2.776110053062439 | -2.2409004497528078 | -1.0223817014694214 | -0.7864253520965576 | -0.6037061452865601 | 0.060917205810546896 | 0.20828267186880112 | 0.48788334012031553 | 0.24300194919109347 | 0.5132518112659454 | 0.7604112458229065 | 0.4461615562438965 | 0.7856936752796173 | 0.928072657585144 | 3.340684480667114 | 3.5573596954345703 | 3.6869903659820555 | -1.116567554473877 | -0.8343653082847595 | -0.6963519287109375 | 8.91455307006836 | 9.098039150238037 | 9.238429145812988 | -1.9138937044143676 | -1.7762314677238464 | -1.5578833723068237 |
| 3 | 10.53053009033203 | 10.692829132080078 | 10.83322998046875 | -3.595937671661377 | -3.1261976957321167 | -2.771820592880249 | 0.08012253761291505 | 0.17132862657308578 | 0.22545420050621032 | 1.2077582454681397 | 1.5327622294425964 | 1.7067881536483764 | 0.07260359317064286 | 0.22691094875335693 | 0.5193048334121704 | 0.2732751202583313 | 0.5990770161151886 | 0.8517063140869141 | 0.794383282661438 | 0.9472034573554993 | 0.9842549133300781 | 2.99753457069397 | 3.175831437110901 | 3.349937763214111 | 1.1364484214782715 | 1.5117439031600952 | 1.6828412771224976 | 10.322906341552734 | 10.469072818756104 | 10.588120422363282 | -0.6779753065109253 | -0.458346426486969 | -0.27817421674728393 |
| 4 | 10.329876174926758 | 10.47231388092041 | 10.58506046295166 | -2.953227691650391 | -2.6161922216415405 | -2.3888746166229247 | -1.540432834625244 | -0.5732389688491821 | -0.2879416954517365 | -1.436713728904724 | -1.0432096719741821 | -0.7430683898925782 | 0.02740598365664482 | 0.11742453277111053 | 0.32760893702507016 | 0.1490445476770401 | 0.3344271630048752 | 0.5359427642822265 | 0.3199243128299713 | 0.4704192280769348 | 0.6171543145179749 | 3.5565563011169434 | 3.655835747718811 | 3.7220150947570803 | -1.5357025957107544 | -1.137833595275879 | -0.8546456933021546 | 10.043120079040527 | 10.18197774887085 | 10.286999282836915 | -0.45858083605766287 | 0.4111205041408539 | 1.0012610626220704 |
| 5 | 10.514813385009766 | 10.60422658920288 | 10.680193977355957 | -3.9610337162017824 | -3.7853821516036987 | -3.342250318527222 | -0.7240598225593566 | -0.46453820168972015 | -0.31559689044952394 | -0.8330999779701233 | -0.379890039563179 | -0.14532580554485322 | 0.021441682279109957 | 0.10100274533033371 | 0.235304456949234 | 0.13084600985050202 | 0.26384831964969635 | 0.49491158723831175 | 0.21373106956481933 | 0.3839689791202545 | 0.6037393116950989 | 3.3228086757659914 | 3.421437978744507 | 3.4789054870605467 | -0.8404275512695313 | -0.3925653100013733 | -0.16153272151947023 | 10.242988204956054 | 10.322557926177979 | 10.391872978210449 | -0.49662286281585694 | -0.3015878200531006 | -0.11881242156028748 |
| 6 | 8.673964958190918 | 8.904021263122559 | 9.069251289367676 | -3.033935775756836 | -2.7565478086471558 | -2.618418855667114 | -2.5305645370483396 | -1.6592541337013245 | -0.9208667135238647 | -1.0269253587722778 | -0.8940448462963104 | -0.7102961874008179 | 0.05037546142935753 | 0.20386016368865967 | 0.46557670950889585 | 0.2620150804519653 | 0.4930070489645004 | 0.7582269859313965 | 0.4829544878005982 | 0.7613232135772705 | 0.9655908298492432 | 3.5014231395721436 | 3.7077839374542236 | 3.8166400718688966 | -1.0213359689712525 | -0.90673828125 | -0.7965976977348328 | 8.420825233459473 | 8.621294498443604 | 8.777588768005371 | -2.1219652271270752 | -2.0492581129074097 | -1.92760910987854 |
| 7 | 8.276817970275879 | 8.46999454498291 | 8.592100143432617 | -3.9012059211730956 | -3.622899293899536 | -3.3821629810333254 | -2.732761068344116 | -1.1937393546104431 | -0.5097035503387451 | -0.7558682584762573 | -0.5702216327190399 | -0.37186248660087584 | 0.04131070226430893 | 0.1624949723482132 | 0.4297352063655853 | 0.20901303112506867 | 0.4375660568475723 | 0.6997544765472412 | 0.3627573871612549 | 0.6439600884914398 | 0.8787775540351868 | 3.076163549423218 | 3.2350751161575317 | 3.3530206966400145 | -0.7532573747634888 | -0.5827709138393402 | -0.3964386796951294 | 8.057681159973145 | 8.22086477279663 | 8.330975189208985 | -2.2304548263549804 | -2.167076826095581 | -1.9705236387252807 |
| 8 | 11.202297058105469 | 11.319639682769775 | 11.424218673706054 | -3.9453994274139403 | -3.7193949222564697 | -3.238027448654175 | 0.014602851495146753 | 0.09616959095001221 | 0.18544672429561612 | 0.2678953278064728 | 0.960591048002243 | 1.6921207523345947 | 0.06151130676269532 | 0.22724857926368713 | 0.49373035311698915 | 0.33778262853622437 | 0.5751250684261322 | 0.7651952981948852 | 0.7106121873855591 | 0.808107316493988 | 0.896643693447113 | 3.0774443244934084 | 3.2303847074508667 | 3.3304523849487304 | 0.32132985472679154 | 0.9549798369407654 | 1.658512315750122 | 10.96874137878418 | 11.069159507751465 | 11.162392845153809 | -0.22055389285087584 | -0.0710608959197998 | 0.06708817481994629 |
| 9 | 9.977440605163574 | 10.094542503356934 | 10.219714813232422 | -3.985952320098877 | -3.9415669441223145 | -3.77304349899292 | -0.36748247146606444 | -0.19631701707839966 | -0.012043787315487865 | -0.4318004941940307 | 0.15985726565122604 | 0.5817402625083923 | 0.04992724120616913 | 0.1546182557940483 | 0.3929023742675781 | 0.1973634272813797 | 0.41424812376499176 | 0.6639959383010864 | 0.38095454454421995 | 0.661058634519577 | 0.8467808771133423 | 3.249329090118408 | 3.3846843242645264 | 3.4813685512542722 | -0.43326798081398005 | 0.1248934455215931 | 0.5391318321228027 | 9.724909133911133 | 9.823789596557617 | 9.936937026977539 | -1.1846681642532348 | -1.0447161197662354 | -0.8543273425102234 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 90 | 8.467938308715821 | 8.612224578857422 | 8.712338600158692 | -3.9118242263793945 | -3.670263648033142 | -3.180136957168579 | -2.9908473682403565 | -2.956583857536316 | -2.8570651149749757 | -0.028349236547946927 | 0.05413089133799076 | 0.11789040118455886 | 0.09046362400054932 | 0.27609992027282715 | 0.5164162445068359 | 0.4397362852096558 | 0.665221631526947 | 0.8431855154037475 | 0.8255544686317444 | 0.9496687948703766 | 0.9840734457969665 | 2.7864186573028564 | 2.9526177644729614 | 3.0897021007537844 | -0.05210614264011383 | 0.047279443591833115 | 0.09815156489610671 | 8.273713340759278 | 8.411657333374023 | 8.497819938659667 | -2.3538817596435546 | -2.34079372882843 | -2.319191074371338 |
| 91 | 9.686736488342286 | 9.733762264251709 | 9.797216911315918 | -3.9658040142059328 | -3.8698363304138184 | -3.7642845249176027 | -2.303443880081177 | -1.4480793476104736 | -0.6218935918807984 | -1.8089547204971312 | -0.656389445066452 | 0.07851898282766337 | 0.1308672147989273 | 0.34924526512622833 | 0.627486412525177 | 0.5130164456367493 | 0.7534611523151398 | 0.8724988341331482 | 0.894643337726593 | 0.9198218286037445 | 0.9480320239067077 | 2.9107672119140626 | 3.0393165349960327 | 3.1747095584869385 | -1.8312068796157837 | -0.6722643375396729 | 0.04612042009830472 | 9.470166969299317 | 9.508735656738281 | 9.559801445007324 | -1.7165182733535767 | -1.653548002243042 | -1.5891746520996093 |
| 92 | 10.217627639770507 | 10.357178688049316 | 10.478487930297852 | -2.5796295261383055 | -2.2853704690933228 | -1.9981221866607666 | 0.222074516415596 | 0.2787896692752838 | 0.3277129065990448 | 1.2711222171783447 | 1.462171733379364 | 1.6259286308288574 | 0.049269839227199554 | 0.16637475788593292 | 0.3933703243732452 | 0.20620704293251038 | 0.43472549319267273 | 0.6896352601051331 | 0.41205934882164 | 0.6421559453010559 | 0.8785081815719604 | 3.151320390701294 | 3.2947826385498047 | 3.401735153198242 | 1.250854549407959 | 1.41789972782135 | 1.5603513288497926 | 10.002862510681153 | 10.124136924743652 | 10.226281471252442 | -0.21461944878101347 | 0.013864066451787949 | 0.25478840589523316 |
| 93 | 8.269257698059082 | 8.421949863433838 | 8.532754821777344 | -3.9800770568847654 | -3.8856383562088013 | -3.686370267868042 | -2.049650526046753 | -1.238416314125061 | -0.5661825037002564 | -1.6835411643981935 | -1.4385753870010376 | -1.2157229423522948 | 0.04319661229848862 | 0.12894362956285477 | 0.3485598075389862 | 0.1681357824802399 | 0.36897099018096924 | 0.6575239300727844 | 0.3060441219806671 | 0.5735911726951599 | 0.8646877455711365 | 3.4178222751617433 | 3.559737205505371 | 3.638593626022339 | -1.6923364114761352 | -1.4618347883224487 | -1.2330515956878663 | 8.005196952819825 | 8.138813972473145 | 8.237160453796387 | -1.9997316598892212 | -1.8996492624282837 | -1.7474086475372315 |
| 94 | 8.870909538269043 | 8.981854438781738 | 9.082789382934571 | -2.7534476566314696 | -2.2134265899658203 | -2.105312871932983 | -0.30668712854385377 | -0.24259769916534424 | -0.1976001250743866 | 0.755060613155365 | 0.8957036733627319 | 0.9989834952354432 | 0.0936733728647232 | 0.31911344826221466 | 0.6306946063041687 | 0.382929128408432 | 0.6956470608711243 | 0.881607608795166 | 0.8286622357368469 | 0.9747115969657898 | 0.9920064449310303 | 2.4485173416137695 | 2.659457802772522 | 2.8260947036743165 | 0.8028233504295349 | 0.9099208116531372 | 0.9815092158317565 | 8.7202099609375 | 8.827516555786133 | 8.914336585998536 | -1.7104293346405028 | -1.6195834279060364 | -1.5417509603500366 |
| 95 | 8.876353797912598 | 9.07252836227417 | 9.26684787750244 | -3.56313009262085 | -3.098684549331665 | -2.753771142959595 | -1.2248799753189086 | -0.6096833050251007 | -0.3049050271511078 | -1.5739735460281372 | -1.1651009917259216 | -0.9516885495185852 | 0.0527811573445797 | 0.20512691885232925 | 0.4391037952899932 | 0.27129032373428347 | 0.5174216032028198 | 0.7708363199234008 | 0.5549591255187988 | 0.8238781988620758 | 0.9430371761322022 | 3.534418153762817 | 3.740338921546936 | 3.8651330947875975 | -1.6079508304595946 | -1.243908405303955 | -1.01923330783844 | 8.604943199157715 | 8.78653335571289 | 8.975468788146973 | -1.7581797599792481 | -1.4381957650184631 | -0.998660900592804 |
| 96 | 10.767762298583984 | 10.862956523895264 | 10.960644073486328 | -2.109587984085083 | -1.8663303852081299 | -1.6872119426727294 | -1.0490061378479003 | -0.46603237092494965 | -0.14617968440055848 | -1.3017631959915161 | -1.0654341578483582 | -0.7424264407157898 | 0.0099351941421628 | 0.0298763457685709 | 0.09583563566207885 | 0.05233375757932663 | 0.11529103294014931 | 0.23290217399597168 | 0.07037654668092728 | 0.13753002136945724 | 0.24647728085517884 | 3.734656085968018 | 3.7705652713775635 | 3.797588701248169 | -1.507627477645874 | -1.29646235704422 | -1.0310893535614014 | 10.468984832763672 | 10.555097579956055 | 10.652542839050293 | -0.44900829553604127 | 0.06662878021597862 | 0.8890600728988647 |
| 97 | 10.841490707397462 | 10.954441547393799 | 11.042369956970214 | -3.7381189346313475 | -3.227435350418091 | -2.8807295989990234 | -0.7122359299659728 | -0.2669187933206558 | -0.12407686084508895 | -1.1265155267715454 | -0.8942262530326843 | -0.5625531268119812 | 0.010951659232378006 | 0.03996937908232212 | 0.09636335015296936 | 0.06103120043873787 | 0.11173447966575623 | 0.20722556591033933 | 0.07074797362089157 | 0.13088052719831467 | 0.24608759105205535 | 3.452436714172363 | 3.4879279136657715 | 3.511270275115967 | -1.1237796783447265 | -0.8831528425216675 | -0.5684370803833009 | 10.555943489074707 | 10.667087078094482 | 10.749487800598144 | 1.2401532793045045 | 1.8353420495986938 | 2.189707489013672 |
| 98 | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan | nan |
| 99 | 10.627729835510253 | 10.742479801177979 | 10.87334373474121 | -1.8682724475860595 | -1.7194148898124695 | -1.6048885536193849 | -1.6869652938842774 | -1.212232232093811 | -0.9270537066459656 | 2.3444686794281004 | 2.4985800981521606 | 2.5920894813537596 | 0.07050890445709229 | 0.23992863297462463 | 0.4818552827835083 | 0.3178158748149872 | 0.6202527582645416 | 0.8510062885284424 | 0.5908016276359558 | 0.8581962585449219 | 0.9689001560211181 | 2.401473731994629 | 2.559554100036621 | 2.7133460330963133 | 2.3901148319244383 | 2.4927940368652344 | 2.5688255882263182 | 10.48086639404297 | 10.582380771636963 | 10.701724815368653 | -2.1085242366790773 | -2.033019185066223 | -1.9606974172592164 |
And we can plot the star-forming main sequence from the fitted catalogue.
[7]:
import matplotlib.pyplot as plt
plt.scatter(post_tab["log_mass_50"], post_tab["log_sfr_50"])
plt.xlabel("Stellar Mass (log M_sun)")
plt.ylabel("SFR (log M_sun/yr)")
[7]:
Text(0, 0.5, 'SFR (log M_sun/yr)')
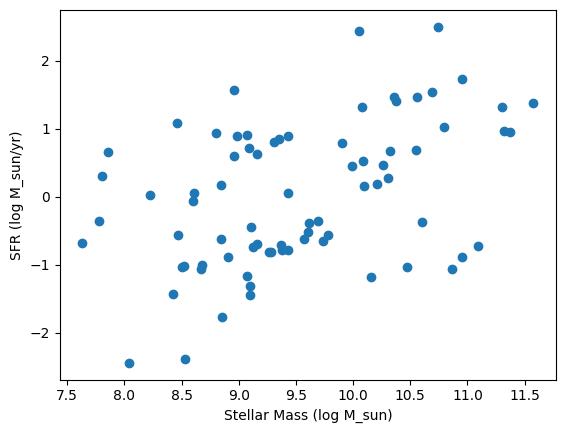
You may notice some of rows and nans. This is because those rows had missing data that we chose not to impute, which can’t be handled by the SBI inference method.
We could set missing_data_mcmc to True to impute those missing values instead.
We can also recover and plot the SEDs, which we’ll do for the first few sources in the catalogue.
[8]:
post_tab, data = fitter.fit_catalogue(
cat[:4],
columns_to_feature_names=conversion_dict,
flux_units=Jy,
check_out_of_distribution=False,
recover_SEDs=True,
plot_SEDs=True,
missing_data_flag=np.nan,
num_samples=300,
append_to_input=False,
)
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
Sampling from posterior: 100%|██████████| 4/4 [00:00<00:00, 18.92it/s]
2025-11-13 20:10:14,203 | synference | INFO | Obtained posterior samples.
Recovering SEDs from posterior samples...: 0%| | 0/4 [00:00<?, ?it/s]
2025-11-13 20:10:14,309 | synference | WARNING | The following parameters are not used by the simulator: ['log10_Av', 'log_sfr', 'sfh_quantile_25', 'sfh_quantile_50', 'sfh_quantile_75', 'log10_mass_weighted_age', 'log10_floor_sfr_10', 'log_surviving_mass', 'beta', 'Av', 'mass_weighted_age', 'floor_sfr_10']
2025-11-13 20:11:09,440 | synference | INFO | Saving SED plot to /opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/models/BPASS_DenseBasis_v4_final/plots//BPASS_DenseBasis_v4_final_SED_0
Recovering SEDs from posterior samples...: 25%|██▌ | 1/4 [00:56<02:49, 56.56s/it]
2025-11-13 20:12:06,258 | synference | INFO | Saving SED plot to /opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/models/BPASS_DenseBasis_v4_final/plots//BPASS_DenseBasis_v4_final_SED_1
Recovering SEDs from posterior samples...: 50%|█████ | 2/4 [01:53<01:52, 56.50s/it]
2025-11-13 20:13:02,653 | synference | INFO | Saving SED plot to /opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/models/BPASS_DenseBasis_v4_final/plots//BPASS_DenseBasis_v4_final_SED_2
Recovering SEDs from posterior samples...: 75%|███████▌ | 3/4 [02:49<00:56, 56.45s/it]
2025-11-13 20:13:56,153 | synference | INFO | Saving SED plot to /opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/models/BPASS_DenseBasis_v4_final/plots//BPASS_DenseBasis_v4_final_SED_3
Recovering SEDs from posterior samples...: 100%|██████████| 4/4 [03:42<00:00, 55.73s/it]
If you recover the SEDs then a nested dictionary with the SED posteriors are also returned. Initial key is the source index or ID, then within that there are keys for ‘wav’, ‘fnu_quantiles’, ‘wav’, ‘phot_wav’, ‘phot_fnu_draws’ and ‘fig’.
[9]:
data[1].keys()
[9]:
dict_keys(['wav', 'fnu_quantiles', 'phot_wav', 'phot_fnu_draws', 'fig'])
Here are the SED plots:
[10]:
for key in data.keys():
display(data[key]["fig"])
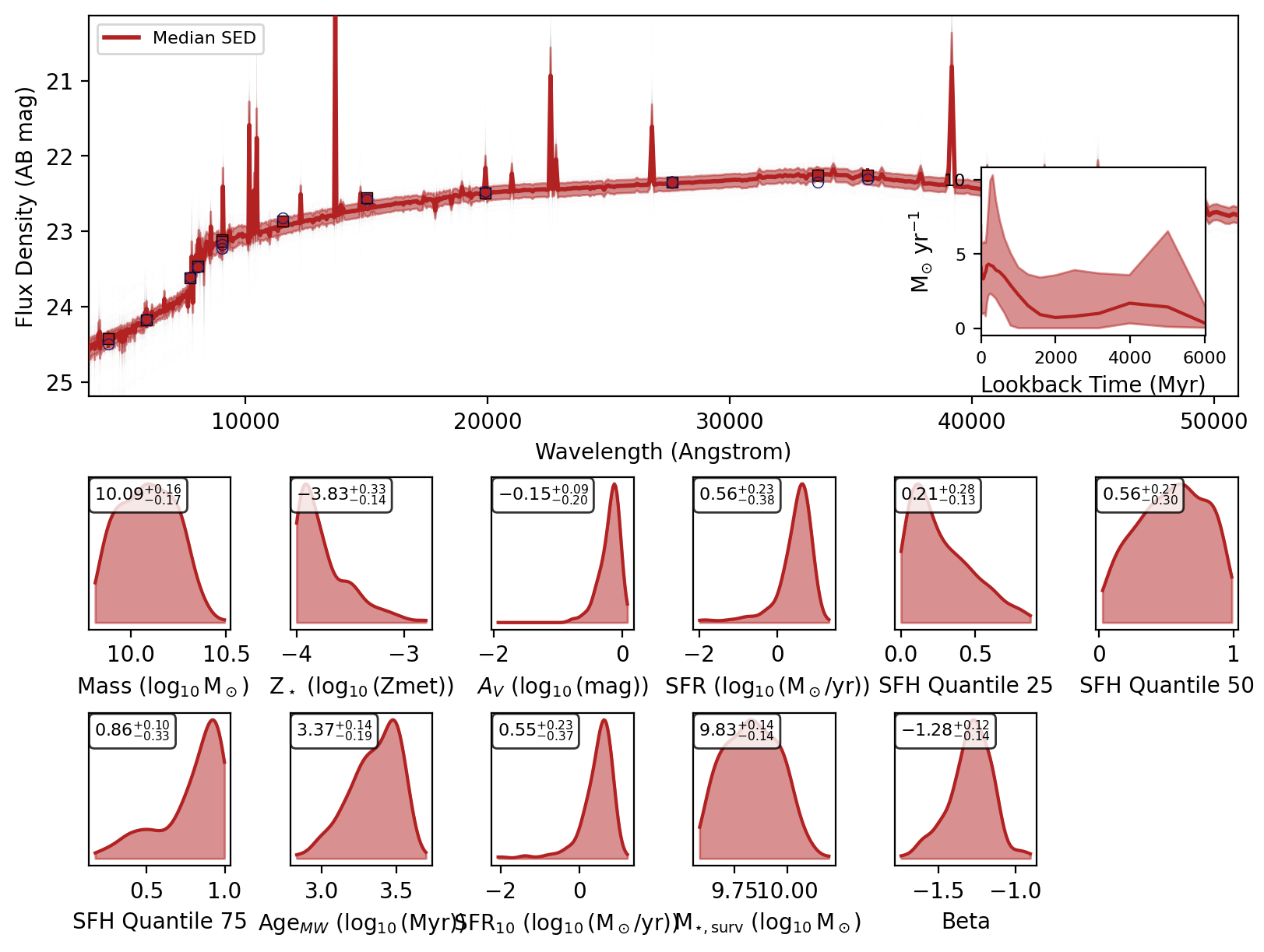
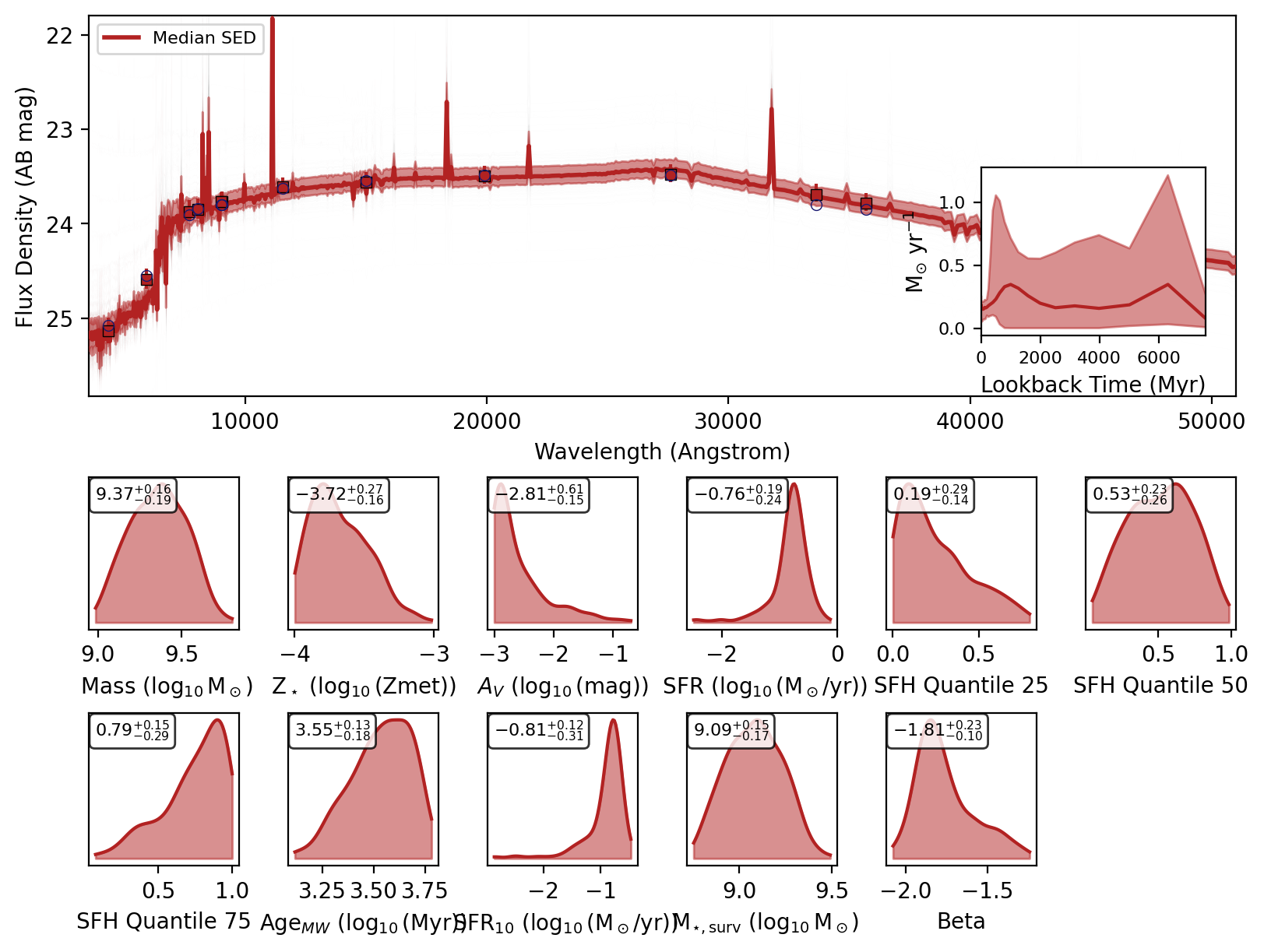
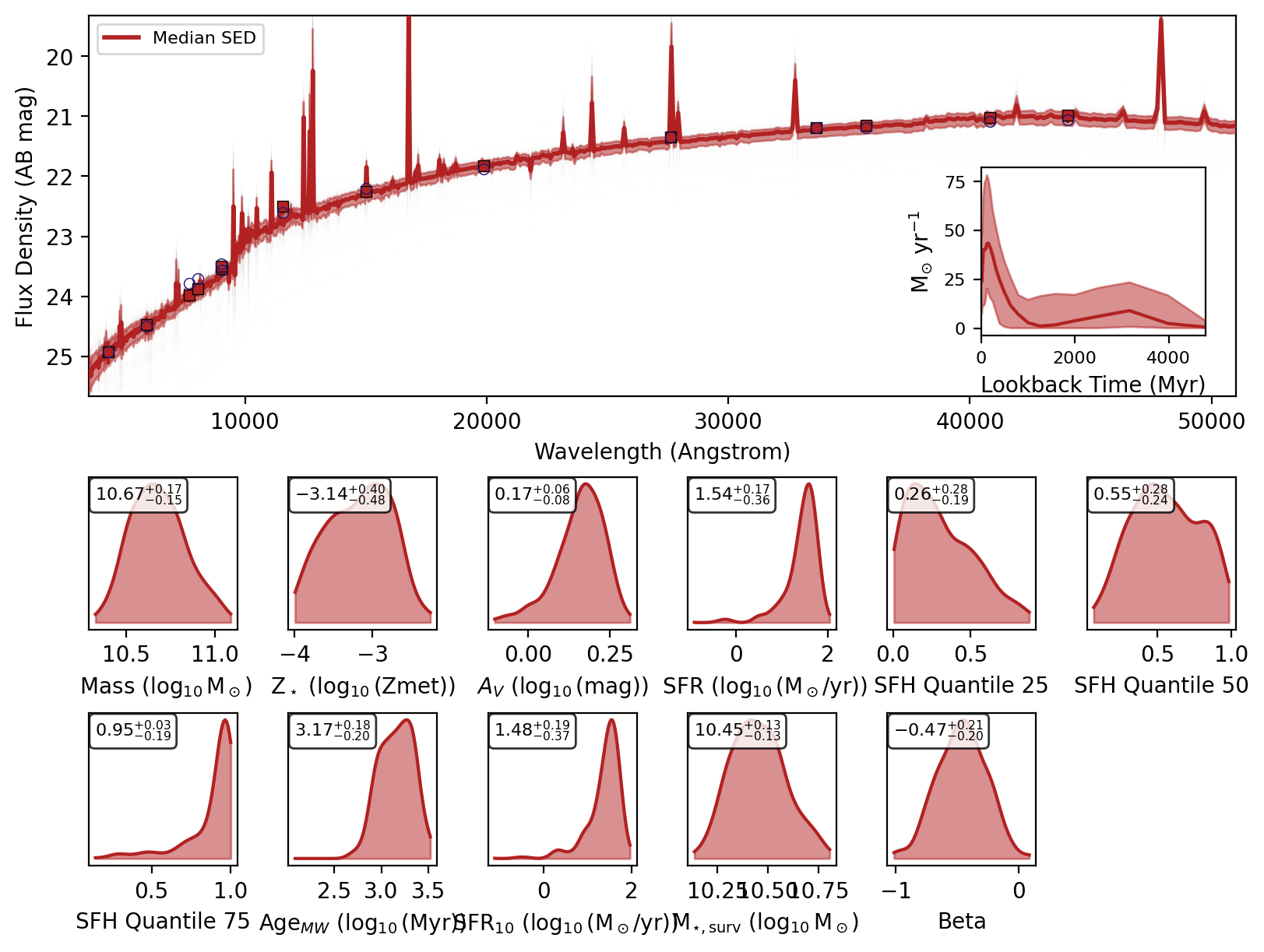
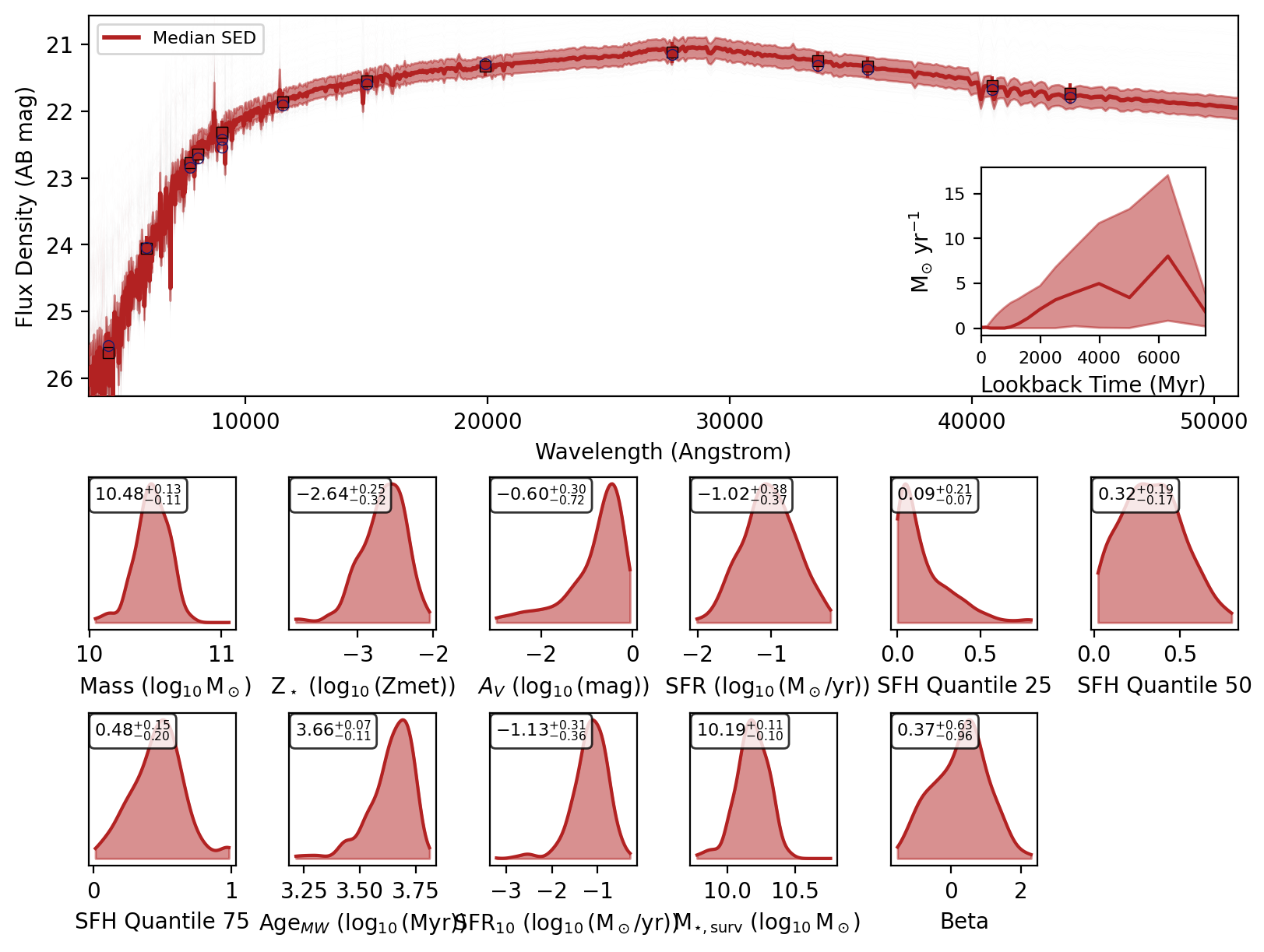
We can check the feature array used for inference by setting return_feature_array=True.
[11]:
feature_array, mask = post_tab, data = fitter.fit_catalogue(
cat,
columns_to_feature_names=conversion_dict,
flux_units=Jy,
check_out_of_distribution=False,
return_feature_array=True,
missing_data_flag=np.nan,
num_samples=300,
append_to_input=False,
)
feature_array
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
WARNING {'true_flux_units': Jy, 'out_units': 'AB'} arguments will have no effect with this model. Input must be in Jy.
2025-11-13 20:14:01,535 | synference | INFO | Removing 54 observations with missing data.
[11]:
array([[2.4493113e+01, 2.4163765e+01, 2.3604200e+01, ..., 9.3294436e-04,
8.9689222e-04, 1.0870000e+00],
[2.5077990e+01, 2.4549706e+01, 2.3904078e+01, ..., 6.5881172e-03,
7.1780500e-03, 6.9384003e-01],
[2.4922672e+01, 2.4493999e+01, 2.3787994e+01, ..., 1.1117980e-03,
8.8765030e-04, 1.5530000e+00],
...,
[2.6450207e+01, 2.4133791e+01, 2.2739664e+01, ..., 3.6288964e-04,
2.9280997e-04, 6.6799998e-01],
[2.7736353e+01, 2.6571774e+01, 2.5916496e+01, ..., 4.7382890e-04,
3.4990878e-04, 1.6912301e+00],
[2.3816370e+01, 2.3439030e+01, 2.3062237e+01, ..., 6.9473550e-04,
6.0235627e-04, 5.7800002e+00]], shape=(76, 29), dtype=float32)
We can also run prior predictive checks to identify outliers, by setting check_out_of_distribution=True. The available list of methods is any of those in PYOD (https://pyod.readthedocs.io/en/latest/). You can set them with the ‘outlier_methods’ argument as a list of strings.